Practical: MACE
In this practical, you’ll explore how to train and evaluate a MACE model. We will use EMT potential for demonstration, practically, you can use DFT or other potentials as well.
Data Generation¶
First, we need to generate the training data. We will use EMT potential running on the ASAP3 package for the Copper system as an example. Then use MACE to train a model on this system. Finally, we will compare our results with the target potential.
Our data will be generated through molecular dynamics with NPT ensemble, at 800 K, and save 40 structures.
from ase.md.velocitydistribution import MaxwellBoltzmannDistribution,Stationary
from ase.md.nvtberendsen import NVTBerendsen
from ase.md.verlet import VelocityVerlet
from ase.md.npt import NPT
from ase.md import MDLogger
from ase import units
from ase.io import write
import os
from time import perf_counter
from ase.calculators.singlepoint import SinglePointCalculator
def run_md(atoms, calculator, ensemble, time_step, temperature, num_md_steps, num_interval, output_filename):
# Set calculator (EMT in this case)
atoms.calc = calculator
# Set the momenta corresponding to the given "temperature"
MaxwellBoltzmannDistribution(atoms, temperature_K=temperature,force_temp=True)
Stationary(atoms) # Set zero total momentum to avoid drifting
# Set output filenames
log_filename = output_filename + ".log"
print("log_filename = ",log_filename)
traj_filename = output_filename + ".traj"
print("traj_filename = ",traj_filename)
xyz_file = output_filename + ".xyz"
print("xyz_filename = ",xyz_file)
# Remove old files if they exist
if os.path.exists(log_filename): os.remove(log_filename)
if os.path.exists(traj_filename): os.remove(traj_filename)
if os.path.exists(xyz_file): os.remove(xyz_file)
# Define the MD dynamics class object
if ensemble == 'nve':
dyn = VelocityVerlet(atoms,
time_step * units.fs,
trajectory = traj_filename,
loginterval=num_interval
)
elif ensemble == 'nvt':
dyn = NVTBerendsen(atoms,
time_step * units.fs,
temperature_K=temperature,
taut=100.0 * units.fs,
trajectory = traj_filename,
loginterval=num_interval
)
elif ensemble == 'npt':
sigma = 1.0 # External pressure in bar
ttime = 20.0 # Time constant in fs
pfactor = 2e6 # Barostat parameter in GPa
dyn = NPT(atoms,
time_step*units.fs,
temperature_K = temperature,
externalstress = sigma*units.bar,
ttime = ttime*units.fs,
pfactor = pfactor*units.GPa*(units.fs**2),
logfile = log_filename,
trajectory = traj_filename,
loginterval=num_interval
)
else:
raise ValueError("Invalid ensemble, must be nve, nvt, or npt")
# Print statements
def print_dyn():
imd = dyn.get_number_of_steps()
etot = atoms.get_total_energy()
temp_K = atoms.get_temperature()
stress = atoms.get_stress(include_ideal_gas=True)/units.GPa
stress_ave = (stress[0]+stress[1]+stress[2])/3.0
elapsed_time = perf_counter() - start_time
print(f" {imd: >3} {etot:.3f} {temp_K:.2f} {stress_ave:.2f} {stress[0]:.2f} {stress[1]:.2f} {stress[2]:.2f} {stress[3]:.2f} {stress[4]:.2f} {stress[5]:.2f} {elapsed_time:.3f}")
def save_xyz():
# Save the current configuration to the trajectory file
atoms_copy = dyn.atoms.copy()
atoms_copy.calc = SinglePointCalculator(atoms_copy,
energy=atoms.get_potential_energy(),
forces = atoms.get_forces(),
stress = atoms.get_stress(include_ideal_gas=True))
# atoms_copy.info['REF_energy']= atoms.get_potential_energy()
# atoms_copy.set_array(forces,'REF_forces')
write(xyz_file, atoms_copy, format='extxyz', append=True)
dyn.attach(print_dyn, interval=num_interval)
dyn.attach(save_xyz, interval=num_interval)
# Set MD logger
dyn.attach(MDLogger(dyn, atoms, log_filename, header=True, stress=True,peratom=True, mode="w"), interval=num_interval)
# Now run MD simulation
print(f" imd Etot(eV) T(K) stress(mean,xx,yy,zz,yz,xz,xy)(GPa) elapsed_time(sec)")
start_time = perf_counter()
dyn.run(num_md_steps)
from asap3 import EMT
from ase.build import bulk
calculator = EMT()
# Set up a fcc-Cu crystal
atoms = bulk("Cu", "fcc", cubic=True, a=3.615)
atoms.pbc = True
atoms *= 2 # 2x2x2 supercell
print("atoms = ",atoms)
# input parameters
time_step = 1.0 # MD step size in fsec
temperature = 800 # Temperature in Kelvin
num_md_steps = 2000 # Total number of MD steps
num_interval = 50 # Print out interval for .log and .traj
output_filename = "./tmp/Cu_fcc_2x2x2"
ensemble = 'npt'
run_md(atoms, calculator, ensemble, time_step, temperature, num_md_steps, num_interval, output_filename)
atoms = Atoms(symbols='Cu32', pbc=True, cell=[7.23, 7.23, 7.23])
log_filename = ./tmp/Cu_fcc_2x2x2.log
traj_filename = ./tmp/Cu_fcc_2x2x2.traj
xyz_filename = ./tmp/Cu_fcc_2x2x2.xyz
imd Etot(eV) T(K) stress(mean,xx,yy,zz,yz,xz,xy)(GPa) elapsed_time(sec)
50 5.711 595.29 -3.28 -3.14 -3.70 -2.99 -1.09 0.13 0.81 0.013
100 11.283 824.59 -8.20 -6.04 -10.23 -8.34 0.20 1.45 0.56 0.025
150 6.508 598.12 2.69 3.53 1.29 3.24 1.38 -0.28 0.09 0.035
200 6.025 701.64 7.84 7.72 8.11 7.69 0.31 -0.06 0.01 0.045
250 7.380 759.06 5.98 6.43 5.83 5.69 0.74 -0.42 0.20 0.054
300 9.632 802.70 -0.08 0.98 -1.64 0.41 0.17 -0.63 0.45 0.063
350 8.280 816.36 -1.04 -0.42 -0.54 -2.16 -0.96 -1.08 -1.40 0.072
400 6.089 770.35 -0.30 -0.10 0.81 -1.59 -1.10 -1.54 -1.31 0.081
450 5.722 704.68 -2.41 -3.46 0.23 -4.00 -1.64 -0.41 -0.63 0.090
500 6.450 733.42 -4.46 -4.99 -3.21 -5.16 -0.10 -0.54 -1.62 0.099
550 7.420 880.43 -4.23 -4.29 -2.99 -5.40 -0.76 -0.27 -0.31 0.108
600 7.215 941.88 -0.80 -1.39 -0.17 -0.83 -0.21 0.65 -1.22 0.117
650 6.132 545.67 1.81 0.77 2.39 2.28 0.62 0.41 0.22 0.126
700 7.341 746.01 3.73 2.16 4.64 4.40 0.47 0.42 1.14 0.135
750 7.276 743.72 3.90 2.86 3.81 5.01 0.52 1.31 1.62 0.144
800 8.639 907.39 1.23 1.41 0.28 2.01 -0.28 0.79 0.48 0.153
850 7.746 850.50 0.57 0.12 0.64 0.94 -0.31 0.83 0.38 0.163
900 6.998 691.57 -1.54 -1.56 -1.86 -1.21 -0.04 -0.66 2.04 0.172
950 6.350 726.59 -1.83 -3.23 -2.32 0.07 1.74 0.89 1.64 0.181
1000 5.953 769.82 -1.81 -3.39 -2.65 0.62 2.12 0.41 2.03 0.190
1050 7.151 811.22 -3.23 -4.26 -3.58 -1.84 2.72 1.00 0.95 0.200
1100 7.789 877.54 -2.17 -2.85 -1.27 -2.40 0.24 -0.46 0.87 0.209
1150 7.234 845.56 1.10 -0.60 1.76 2.16 1.02 -0.87 -0.89 0.219
1200 6.761 696.99 3.21 2.00 2.81 4.82 0.28 0.31 -1.28 0.229
1250 7.129 763.99 3.74 3.49 3.77 3.97 -0.29 -1.10 -1.40 0.238
1300 8.144 899.20 1.96 1.97 3.24 0.68 -0.79 -0.25 -2.23 0.246
1350 7.224 731.25 0.08 0.23 -0.43 0.45 0.02 -0.45 -1.70 0.255
1400 6.090 761.29 0.06 1.86 -1.22 -0.44 -0.93 -0.76 -1.15 0.264
1450 6.997 952.12 -2.27 -0.45 -3.15 -3.21 -1.61 -0.94 -1.59 0.273
1500 5.822 613.52 -3.10 -0.92 -4.28 -4.10 -1.18 -1.06 -0.73 0.282
1550 6.586 737.82 -3.03 -0.54 -4.19 -4.36 -1.58 -0.63 -1.10 0.291
1600 8.030 1050.62 -1.66 1.02 -2.00 -3.99 -1.73 -1.31 -0.03 0.300
1650 6.054 702.51 2.14 3.67 1.75 1.00 -0.51 -0.32 0.30 0.309
1700 6.768 607.74 2.07 3.79 1.59 0.83 -1.31 0.18 -0.21 0.318
1750 7.701 835.01 2.88 4.81 3.01 0.83 0.47 0.88 0.57 0.326
1800 7.092 753.19 2.29 2.87 2.52 1.49 0.50 0.25 0.32 0.335
1850 7.195 737.18 0.10 -0.14 0.13 0.31 0.27 1.40 0.42 0.343
1900 6.650 815.18 -0.33 0.64 -0.44 -1.20 0.75 0.77 1.77 0.352
1950 6.066 596.18 -2.43 -2.67 -1.29 -3.34 1.90 1.49 2.06 0.361
2000 6.869 790.03 -3.46 -4.52 -3.64 -2.21 0.41 1.85 1.97 0.370
Compute E0¶
Atomic energies are computed using the EMT potential by putting a single atom in a box. This is essential for the MACE model.
from ase import Atoms
# Create a single Cu atom in a large box (to simulate vacuum conditions)
single_Cu = Atoms('Cu', positions=[[0, 0, 0]], cell=[20, 20, 20], pbc=False)
# Use the already defined calculator (EMT)
single_Cu.calc = calculator
# Compute and print the atomic energy
energy = single_Cu.get_potential_energy()
print("Atomic energy of single Cu atom: {:.4f} eV".format(energy))Atomic energy of single Cu atom: 3.5100 eV
import pandas as pd
log_filename = output_filename + ".log"
df = pd.read_csv(log_filename, delim_whitespace=True, skiprows=1,
names=['Time[ps]','Etot/N[eV]','Epot/N[eV]','Ekin/N[eV]','T[K]','stress(xx)','stress(yy)','stress(zz)','stress(yz)','stress(xz)','stress(xy)'])
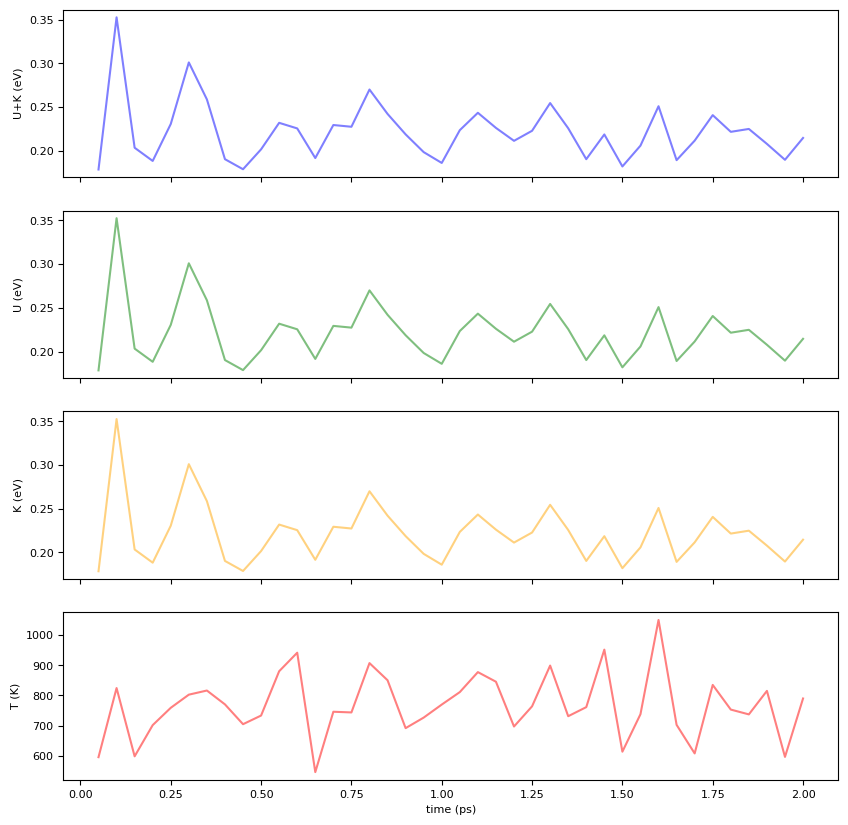
dfimport matplotlib.pyplot as plt
fig = plt.figure(figsize=(10, 10))
#color = 'tab:grey'
ax1 = fig.add_subplot(4, 1, 1)
ax1.set_xticklabels([])
ax1.set_ylabel('U+K (eV)')
ax1.plot(df["Time[ps]"], df["Etot/N[eV]"], color="blue",alpha=0.5)
ax2 = fig.add_subplot(4, 1, 2)
ax2.set_xticklabels([])
ax2.set_ylabel('U (eV)')
ax2.plot(df["Time[ps]"], df["Etot/N[eV]"], color="green",alpha=0.5)
ax3 = fig.add_subplot(4, 1, 3)
ax3.set_xticklabels([])
ax3.set_ylabel('K (eV)')
ax3.plot(df["Time[ps]"], df["Etot/N[eV]"], color="orange",alpha=0.5)
ax4 = fig.add_subplot(4, 1, 4)
ax4.set_xlabel('time (ps)')
ax4.set_ylabel('T (K)')
ax4.plot(df["Time[ps]"], df["T[K]"], color="red",alpha=0.5)
plt.show()
Data Splitting¶
We then need to devide the data into training/validation and test sets. The training set is used to train the MACE model, while the test set is used to evaluate its performance. We will use a 90-10 split for this example.
from ase.io import read, write
import random
atoms = read('./tmp/Cu_fcc_2x2x2.xyz', index=":", format='extxyz',parallel=True)
print(f"Number of data: {len(atoms)}")
# Randomly shuffle the data
seed = 123
random.seed(seed)
# determine the index to split an 90% / 10% data
split_idx = int(0.9 * len(atoms))
indices = list(range(len(atoms)))
random.shuffle(indices)
train_atoms = [atoms[i] for i in indices[:split_idx]]
test_atoms = [atoms[i] for i in indices[split_idx:]]
write("tmp/train_valid.xyz", train_atoms, format="extxyz")
write("tmp/test.xyz", test_atoms, format="extxyz")Number of data: 40
Training Configuration¶
You can use the code below to generate training configuration. For more information, please refer to: https://
%%writefile ./tmp/training_config.yml
model: "MACE"
num_channels: 32
max_L: 0
r_max: 4.0
name: "mace01"
model_dir: "tmp/MACE_models"
log_dir: "tmp/MACE_models"
checkpoints_dir: "tmp/MACE_models"
results_dir: "tmp/MACE_models"
train_file: "tmp/train_valid.xyz"
valid_fraction: 0.10
test_file: "tmp/test.xyz"
energy_key: "energy"
forces_key: "forces"
stress_key: "stress"
E0s: "{29:3.5100}"
device: cpu
batch_size: 4
max_num_epochs: 50
swa: True
seed: 123
Overwriting ./tmp/training_config.yml
import warnings
warnings.filterwarnings("ignore")
from mace.cli.run_train import main as mace_run_train_main
import sys
import logging
def train_mace(config_file_path):
logging.getLogger().handlers.clear()
sys.argv = ["program", "--config", config_file_path]
mace_run_train_main()
Training the MACE Model¶
Now we can start training the MACE model. This process might be time-consuming, depending on the size of your dataset and the complexity of the model. The training process involves optimizing the model parameters to minimize the difference between the predicted and target energies and forces.
train_mace("tmp/training_config.yml")2025-04-16 03:48:29.777 INFO: ===========VERIFYING SETTINGS===========
2025-04-16 03:48:29.778 INFO: MACE version: 0.3.12
2025-04-16 03:48:29.778 INFO: Using CPU
2025-04-16 03:48:29.849 INFO: ===========LOADING INPUT DATA===========
2025-04-16 03:48:29.850 INFO: Using heads: ['default']
2025-04-16 03:48:29.851 INFO: ============= Processing head default ===========
2025-04-16 03:48:29.867 WARNING: Since ASE version 3.23.0b1, using energy_key 'energy' is no longer safe when communicating between MACE and ASE. We recommend using a different key, rewriting 'energy' to 'REF_energy'. You need to use --energy_key='REF_energy' to specify the chosen key name.
2025-04-16 03:48:29.870 WARNING: Since ASE version 3.23.0b1, using forces_key 'forces' is no longer safe when communicating between MACE and ASE. We recommend using a different key, rewriting 'forces' to 'REF_forces'. You need to use --forces_key='REF_forces' to specify the chosen key name.
2025-04-16 03:48:29.873 WARNING: Since ASE version 3.23.0b1, using stress_key 'stress' is no longer safe when communicating between MACE and ASE. We recommend using a different key, rewriting 'stress' to 'REF_stress'. You need to use --stress_key='REF_stress' to specify the chosen key name.
2025-04-16 03:48:29.877 INFO: Training file 1/1 [36 configs, 36 energy, 36 forces, 36 stresses] loaded from 'tmp/train_valid.xyz'
2025-04-16 03:48:29.878 INFO: Total training set [36 configs, 36 energy, 36 forces, 36 stresses]
2025-04-16 03:48:29.878 INFO: Using random 10% of training set for validation with following indices: [27, 25, 13]
2025-04-16 03:48:29.879 INFO: Validation set contains 3 configurations [3 energy, 3 forces, 3 stresses]
2025-04-16 03:48:29.884 WARNING: Since ASE version 3.23.0b1, using energy_key 'energy' is no longer safe when communicating between MACE and ASE. We recommend using a different key, rewriting 'energy' to 'REF_energy'. You need to use --energy_key='REF_energy' to specify the chosen key name.
2025-04-16 03:48:29.885 WARNING: Since ASE version 3.23.0b1, using forces_key 'forces' is no longer safe when communicating between MACE and ASE. We recommend using a different key, rewriting 'forces' to 'REF_forces'. You need to use --forces_key='REF_forces' to specify the chosen key name.
2025-04-16 03:48:29.886 WARNING: Since ASE version 3.23.0b1, using stress_key 'stress' is no longer safe when communicating between MACE and ASE. We recommend using a different key, rewriting 'stress' to 'REF_stress'. You need to use --stress_key='REF_stress' to specify the chosen key name.
2025-04-16 03:48:29.887 INFO: Test file 1/1 [4 configs, 4 energy, 4 forces, 4 stresses] loaded from 'tmp/test.xyz'
2025-04-16 03:48:29.888 INFO: Total test set (4 configs):
2025-04-16 03:48:29.888 INFO: Default_default: 4 configs, 4 energy, 4 forces, 4 stresses
2025-04-16 03:48:29.889 INFO: Total number of configurations: train=33, valid=3, tests=[Default_default: 4],
2025-04-16 03:48:29.893 WARNING: Validation batch size (10) is larger than the number of validation data (3)
2025-04-16 03:48:29.893 INFO: Atomic Numbers used: [29]
2025-04-16 03:48:29.894 INFO: Isolated Atomic Energies (E0s) not in training file, using command line argument
2025-04-16 03:48:29.895 INFO: Atomic Energies used (z: eV) for head default: {29: 3.51}
2025-04-16 03:48:29.895 INFO: Processing datasets for head 'default'
2025-04-16 03:48:29.962 INFO: Combining 1 list datasets for head 'default'
2025-04-16 03:48:29.965 INFO: Head 'default' training dataset size: 33
2025-04-16 03:48:29.966 INFO: Computing average number of neighbors
2025-04-16 03:48:29.973 INFO: Average number of neighbors: 17.7578125
2025-04-16 03:48:29.973 INFO: During training the following quantities will be reported: energy, forces
2025-04-16 03:48:29.974 INFO: ===========MODEL DETAILS===========
2025-04-16 03:48:29.980 INFO: Building model
2025-04-16 03:48:29.981 INFO: Message passing with 32 channels and max_L=0 (32x0e)
2025-04-16 03:48:29.982 INFO: 2 layers, each with correlation order: 3 (body order: 4) and spherical harmonics up to: l=3
2025-04-16 03:48:29.982 INFO: 8 radial and 5 basis functions
2025-04-16 03:48:29.983 INFO: Radial cutoff: 4.0 A (total receptive field for each atom: 8.0 A)
2025-04-16 03:48:29.984 INFO: Distance transform for radial basis functions: None
2025-04-16 03:48:29.985 INFO: Hidden irreps: 32x0e
2025-04-16 03:48:30.543 INFO: Total number of parameters: 53584
2025-04-16 03:48:30.543 INFO:
2025-04-16 03:48:30.544 INFO: ===========OPTIMIZER INFORMATION===========
2025-04-16 03:48:30.545 INFO: Using ADAM as parameter optimizer
2025-04-16 03:48:30.545 INFO: Batch size: 4
2025-04-16 03:48:30.545 INFO: Number of gradient updates: 412
2025-04-16 03:48:30.546 INFO: Learning rate: 0.01, weight decay: 5e-07
2025-04-16 03:48:30.546 INFO: WeightedEnergyForcesLoss(energy_weight=1.000, forces_weight=100.000)
2025-04-16 03:48:30.547 INFO: Stage Two (after 36 epochs) with loss function: WeightedEnergyForcesLoss(energy_weight=1000.000, forces_weight=100.000), with energy weight : 1000.0, forces weight : 100.0 and learning rate : 0.001
2025-04-16 03:48:30.598 INFO: Using gradient clipping with tolerance=10.000
2025-04-16 03:48:30.599 INFO:
2025-04-16 03:48:30.599 INFO: ===========TRAINING===========
2025-04-16 03:48:30.600 INFO: Started training, reporting errors on validation set
2025-04-16 03:48:30.600 INFO: Loss metrics on validation set
2025-04-16 03:48:30.679 INFO: Initial: head: default, loss=19.75021945, RMSE_E_per_atom= 3442.74 meV, RMSE_F= 688.46 meV / A
2025-04-16 03:48:31.930 INFO: Epoch 0: head: default, loss=12.65267467, RMSE_E_per_atom= 3028.92 meV, RMSE_F= 536.50 meV / A
2025-04-16 03:48:33.177 INFO: Epoch 1: head: default, loss=5.00620433, RMSE_E_per_atom= 3025.40 meV, RMSE_F= 242.19 meV / A
2025-04-16 03:48:34.440 INFO: Epoch 2: head: default, loss=4.23638255, RMSE_E_per_atom= 2685.92 meV, RMSE_F= 234.41 meV / A
2025-04-16 03:48:35.765 INFO: Epoch 3: head: default, loss=2.51651391, RMSE_E_per_atom= 2548.65 meV, RMSE_F= 102.66 meV / A
2025-04-16 03:48:36.994 INFO: Epoch 4: head: default, loss=1.67258795, RMSE_E_per_atom= 2071.25 meV, RMSE_F= 85.30 meV / A
2025-04-16 03:48:38.207 INFO: Epoch 5: head: default, loss=0.65792243, RMSE_E_per_atom= 1117.16 meV, RMSE_F= 85.19 meV / A
2025-04-16 03:48:39.770 INFO: Epoch 6: head: default, loss=0.25720635, RMSE_E_per_atom= 186.23 meV, RMSE_F= 85.85 meV / A
2025-04-16 03:48:41.167 INFO: Epoch 7: head: default, loss=0.17780661, RMSE_E_per_atom= 66.33 meV, RMSE_F= 72.73 meV / A
2025-04-16 03:48:42.440 INFO: Epoch 8: head: default, loss=0.11830597, RMSE_E_per_atom= 55.59 meV, RMSE_F= 59.32 meV / A
2025-04-16 03:48:43.671 INFO: Epoch 9: head: default, loss=0.12779799, RMSE_E_per_atom= 217.82 meV, RMSE_F= 57.96 meV / A
2025-04-16 03:48:44.965 INFO: Epoch 10: head: default, loss=0.27851048, RMSE_E_per_atom= 47.46 meV, RMSE_F= 91.28 meV / A
2025-04-16 03:48:46.292 INFO: Epoch 11: head: default, loss=0.33426494, RMSE_E_per_atom= 56.11 meV, RMSE_F= 99.98 meV / A
2025-04-16 03:48:47.530 INFO: Epoch 12: head: default, loss=0.07611942, RMSE_E_per_atom= 17.80 meV, RMSE_F= 47.75 meV / A
2025-04-16 03:48:48.866 INFO: Epoch 13: head: default, loss=0.04599848, RMSE_E_per_atom= 23.20 meV, RMSE_F= 37.08 meV / A
2025-04-16 03:48:50.237 INFO: Epoch 14: head: default, loss=0.07709748, RMSE_E_per_atom= 22.26 meV, RMSE_F= 48.04 meV / A
2025-04-16 03:48:51.480 INFO: Epoch 15: head: default, loss=0.04451550, RMSE_E_per_atom= 29.31 meV, RMSE_F= 36.43 meV / A
2025-04-16 03:48:52.661 INFO: Epoch 16: head: default, loss=0.04861449, RMSE_E_per_atom= 27.74 meV, RMSE_F= 38.09 meV / A
2025-04-16 03:48:53.987 INFO: Epoch 17: head: default, loss=0.02026954, RMSE_E_per_atom= 19.63 meV, RMSE_F= 24.58 meV / A
2025-04-16 03:48:55.292 INFO: Epoch 18: head: default, loss=0.03726097, RMSE_E_per_atom= 12.09 meV, RMSE_F= 33.41 meV / A
2025-04-16 03:48:56.542 INFO: Epoch 19: head: default, loss=0.01654074, RMSE_E_per_atom= 12.63 meV, RMSE_F= 22.24 meV / A
2025-04-16 03:48:57.976 INFO: Epoch 20: head: default, loss=0.01975056, RMSE_E_per_atom= 13.87 meV, RMSE_F= 24.30 meV / A
2025-04-16 03:48:59.240 INFO: Epoch 21: head: default, loss=0.01652776, RMSE_E_per_atom= 32.27 meV, RMSE_F= 22.03 meV / A
2025-04-16 03:49:00.471 INFO: Epoch 22: head: default, loss=0.02347430, RMSE_E_per_atom= 15.22 meV, RMSE_F= 26.49 meV / A
2025-04-16 03:49:01.817 INFO: Epoch 23: head: default, loss=0.01546703, RMSE_E_per_atom= 11.17 meV, RMSE_F= 21.51 meV / A
2025-04-16 03:49:03.165 INFO: Epoch 24: head: default, loss=0.01725255, RMSE_E_per_atom= 11.34 meV, RMSE_F= 22.72 meV / A
2025-04-16 03:49:04.426 INFO: Epoch 25: head: default, loss=0.02783829, RMSE_E_per_atom= 38.14 meV, RMSE_F= 28.65 meV / A
2025-04-16 03:49:05.731 INFO: Epoch 26: head: default, loss=0.01551675, RMSE_E_per_atom= 19.86 meV, RMSE_F= 21.48 meV / A
2025-04-16 03:49:07.050 INFO: Epoch 27: head: default, loss=0.01771259, RMSE_E_per_atom= 12.89 meV, RMSE_F= 23.02 meV / A
2025-04-16 03:49:08.344 INFO: Epoch 28: head: default, loss=0.03725313, RMSE_E_per_atom= 20.50 meV, RMSE_F= 33.37 meV / A
2025-04-16 03:49:09.670 INFO: Epoch 29: head: default, loss=0.02399820, RMSE_E_per_atom= 19.13 meV, RMSE_F= 26.76 meV / A
2025-04-16 03:49:10.906 INFO: Epoch 30: head: default, loss=0.02265691, RMSE_E_per_atom= 15.35 meV, RMSE_F= 26.03 meV / A
2025-04-16 03:49:12.081 INFO: Epoch 31: head: default, loss=0.01256153, RMSE_E_per_atom= 33.81 meV, RMSE_F= 19.12 meV / A
2025-04-16 03:49:13.292 INFO: Epoch 32: head: default, loss=0.01721157, RMSE_E_per_atom= 16.74 meV, RMSE_F= 22.66 meV / A
2025-04-16 03:49:14.512 INFO: Epoch 33: head: default, loss=0.01854147, RMSE_E_per_atom= 10.98 meV, RMSE_F= 23.56 meV / A
2025-04-16 03:49:15.727 INFO: Epoch 34: head: default, loss=0.01330458, RMSE_E_per_atom= 12.42 meV, RMSE_F= 19.94 meV / A
2025-04-16 03:49:16.863 INFO: Epoch 35: head: default, loss=0.01104176, RMSE_E_per_atom= 13.66 meV, RMSE_F= 18.15 meV / A
2025-04-16 03:49:16.900 INFO: Changing loss based on Stage Two Weights
2025-04-16 03:49:18.063 INFO: Epoch 36: head: default, loss=1.18592731, RMSE_E_per_atom= 59.14 meV, RMSE_F= 24.46 meV / A
2025-04-16 03:49:19.278 INFO: Epoch 37: head: default, loss=0.53699927, RMSE_E_per_atom= 39.03 meV, RMSE_F= 29.66 meV / A
2025-04-16 03:49:20.473 INFO: Epoch 38: head: default, loss=0.24354356, RMSE_E_per_atom= 24.81 meV, RMSE_F= 33.96 meV / A
2025-04-16 03:49:21.677 INFO: Epoch 39: head: default, loss=0.16964108, RMSE_E_per_atom= 19.63 meV, RMSE_F= 35.17 meV / A
2025-04-16 03:49:22.914 INFO: Epoch 40: head: default, loss=0.26512949, RMSE_E_per_atom= 26.78 meV, RMSE_F= 27.96 meV / A
2025-04-16 03:49:24.087 INFO: Epoch 41: head: default, loss=0.05382714, RMSE_E_per_atom= 9.79 meV, RMSE_F= 25.61 meV / A
2025-04-16 03:49:25.337 INFO: Epoch 42: head: default, loss=0.25393172, RMSE_E_per_atom= 26.37 meV, RMSE_F= 25.81 meV / A
2025-04-16 03:49:26.576 INFO: Epoch 43: head: default, loss=0.04461799, RMSE_E_per_atom= 8.90 meV, RMSE_F= 23.38 meV / A
2025-04-16 03:49:27.788 INFO: Epoch 44: head: default, loss=0.05101512, RMSE_E_per_atom= 9.99 meV, RMSE_F= 23.09 meV / A
2025-04-16 03:49:28.966 INFO: Epoch 45: head: default, loss=0.14435939, RMSE_E_per_atom= 19.17 meV, RMSE_F= 25.59 meV / A
2025-04-16 03:49:30.173 INFO: Epoch 46: head: default, loss=0.04314693, RMSE_E_per_atom= 8.42 meV, RMSE_F= 24.21 meV / A
2025-04-16 03:49:31.368 INFO: Epoch 47: head: default, loss=0.03901319, RMSE_E_per_atom= 8.24 meV, RMSE_F= 22.19 meV / A
2025-04-16 03:49:32.550 INFO: Epoch 48: head: default, loss=0.06649804, RMSE_E_per_atom= 12.37 meV, RMSE_F= 21.54 meV / A
2025-04-16 03:49:33.689 INFO: Epoch 49: head: default, loss=0.04361523, RMSE_E_per_atom= 8.92 meV, RMSE_F= 22.63 meV / A
2025-04-16 03:49:33.690 INFO: Training complete
2025-04-16 03:49:33.690 INFO:
2025-04-16 03:49:33.690 INFO: ===========RESULTS===========
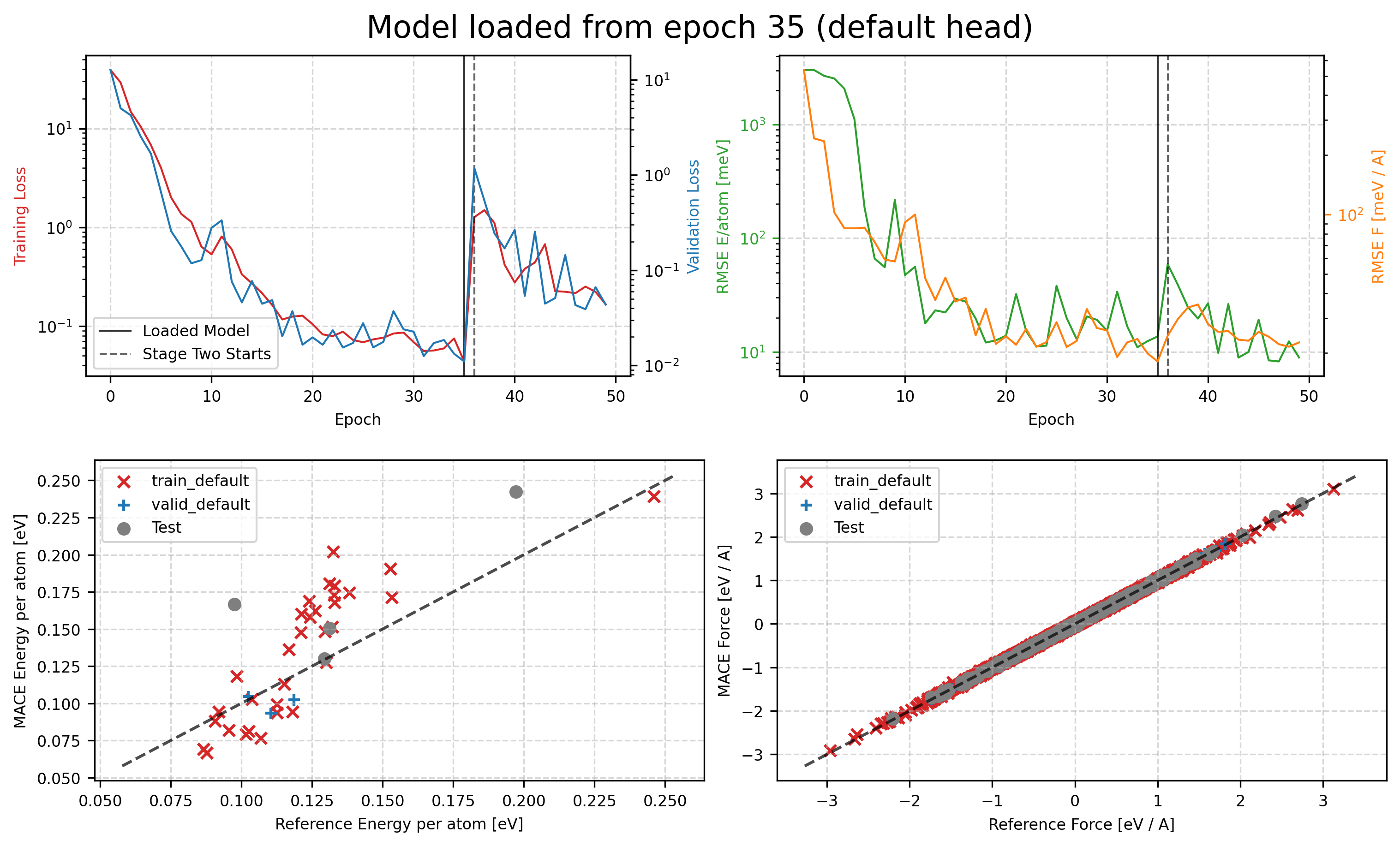
2025-04-16 03:49:33.695 INFO: Loading checkpoint: tmp/MACE_models/mace01_run-123_epoch-35.pt
2025-04-16 03:49:33.712 INFO: Loaded Stage one model from epoch 35 for evaluation
2025-04-16 03:49:33.714 INFO: Saving model to tmp/MACE_models/mace01_run-123.model
2025-04-16 03:49:33.825 INFO: Compiling model, saving metadata to tmp/MACE_models/mace01_compiled.model
2025-04-16 03:49:34.153 INFO: Computing metrics for training, validation, and test sets
2025-04-16 03:49:34.154 INFO: Evaluating train_default ...
2025-04-16 03:49:34.706 INFO: Evaluating valid_default ...
2025-04-16 03:49:34.763 INFO: Error-table on TRAIN and VALID:
+---------------+---------------------+------------------+-------------------+
| config_type | RMSE E / meV / atom | RMSE F / meV / A | relative F RMSE % |
+---------------+---------------------+------------------+-------------------+
| train_default | 29.2 | 19.5 | 2.73 |
| valid_default | 13.7 | 18.1 | 2.65 |
+---------------+---------------------+------------------+-------------------+
2025-04-16 03:49:34.763 INFO: Evaluating Default_default ...
2025-04-16 03:49:34.837 INFO: Error-table on TEST:
+-----------------+---------------------+------------------+-------------------+
| config_type | RMSE E / meV / atom | RMSE F / meV / A | relative F RMSE % |
+-----------------+---------------------+------------------+-------------------+
| Default_default | 42.4 | 20.1 | 2.86 |
+-----------------+---------------------+------------------+-------------------+
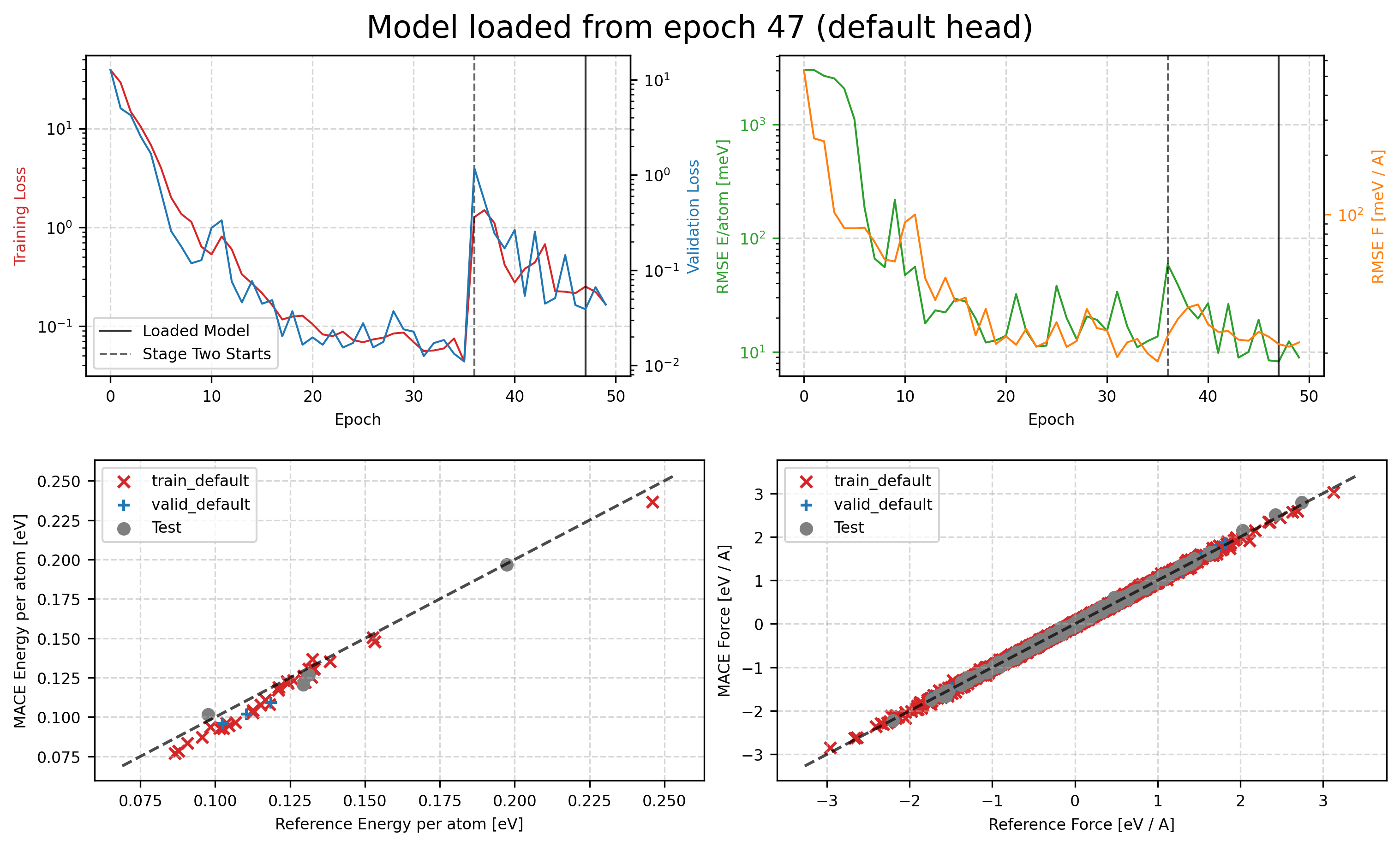
2025-04-16 03:49:36.270 INFO: Loading checkpoint: tmp/MACE_models/mace01_run-123_epoch-47_swa.pt
2025-04-16 03:49:36.275 INFO: Loaded Stage two model from epoch 47 for evaluation
2025-04-16 03:49:36.276 INFO: Saving model to tmp/MACE_models/mace01_run-123_stagetwo.model
2025-04-16 03:49:36.405 INFO: Compiling model, saving metadata tmp/MACE_models/mace01_stagetwo_compiled.model
2025-04-16 03:49:36.755 INFO: Computing metrics for training, validation, and test sets
2025-04-16 03:49:36.756 INFO: Evaluating train_default ...
2025-04-16 03:49:37.184 INFO: Evaluating valid_default ...
2025-04-16 03:49:37.230 INFO: Error-table on TRAIN and VALID:
+---------------+---------------------+------------------+-------------------+
| config_type | RMSE E / meV / atom | RMSE F / meV / A | relative F RMSE % |
+---------------+---------------------+------------------+-------------------+
| train_default | 6.7 | 26.9 | 3.78 |
| valid_default | 8.2 | 22.2 | 3.24 |
+---------------+---------------------+------------------+-------------------+
2025-04-16 03:49:37.230 INFO: Evaluating Default_default ...
2025-04-16 03:49:37.280 INFO: Error-table on TEST:
+-----------------+---------------------+------------------+-------------------+
| config_type | RMSE E / meV / atom | RMSE F / meV / A | relative F RMSE % |
+-----------------+---------------------+------------------+-------------------+
| Default_default | 5.3 | 27.6 | 3.93 |
+-----------------+---------------------+------------------+-------------------+
2025-04-16 03:49:38.376 INFO: Done
Track Training Results¶
You can then check the training curves in tmp/MACE_models/mace01_run-123_train_default_stage_one.png and tmp/MACE_models/mace01_run-123_train_default_stage_two.png to see how the training process goes. The first one is the training curve for stage one, and the second one is for stage two.
from IPython.display import Image, display
display(Image(filename='tmp/MACE_models/mace01_run-123_train_default_stage_one.png'))
display(Image(filename='tmp/MACE_models/mace01_run-123_train_default_stage_two.png'))
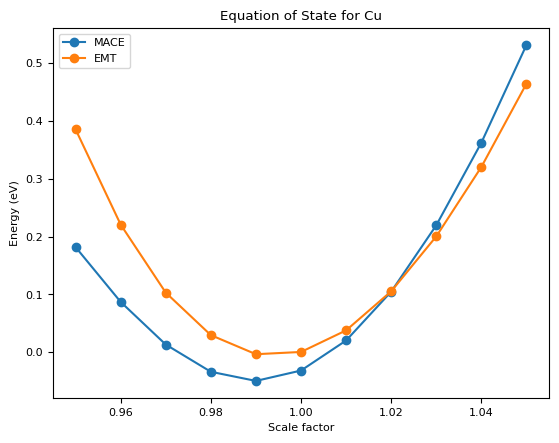
Further Test: Equation of State¶
Then we can test the equation of state of the model.
from mace.calculators import MACECalculator
import numpy as np
from ase.eos import EquationOfState
calculator = MACECalculator(
model_paths="tmp/MACE_models/mace01_run-123_stagetwo.model",
device="cpu")
import matplotlib.pyplot as plt
# Define a range of scale factors
scale_factors = np.linspace(0.95, 1.05, 11)
energies_mace = []
energies_emt = []
volumes = []
atoms = bulk("Cu", "fcc", cubic=True, a=3.615)
atoms.pbc = True
for s in scale_factors:
# Make a copy of the original bulk Cu structure
atoms_scaled = atoms.copy()
# Scale the cell and positions accordingly
atoms_scaled.set_cell(atoms_scaled.get_cell() * s, scale_atoms=True)
atoms_scaled_emt = atoms_scaled.copy()
# Assign the MACE calculator
atoms_scaled.calc = calculator
atoms_scaled_emt.calc = EMT()
volumes.append(atoms_scaled.get_volume())
# Compute and store the potential energy
energies_mace.append(atoms_scaled.get_potential_energy())
energies_emt.append(atoms_scaled_emt.get_potential_energy())
eos_mace = EquationOfState(volumes, energies_mace)
eos_emt = EquationOfState(volumes, energies_emt)
# Fit the EOS to the MACE energies
v0_mace, e0_mace, B_mace = eos_mace.fit()
# Fit the EOS to the EMT energies
v0_emt, e0_emt, B_emt = eos_emt.fit()
print(f"MACE: v0 = {v0_mace:.4f}, e0 = {e0_mace:.4f}, B = {B_mace:.4f}")
print(f"EMT: v0 = {v0_emt:.4f}, e0 = {e0_emt:.4f}, B = {B_emt:.4f}")
# Plot the Equation of State (EOS)
plt.figure()
plt.plot(scale_factors, energies_mace, marker="o", label="MACE")
plt.plot(scale_factors, energies_emt, marker="o", label="EMT")
plt.legend()
plt.xlabel("Scale factor")
plt.ylabel("Energy (eV)")
plt.title("Equation of State for Cu")
plt.show()2025-04-16 03:50:10.278 INFO: Using CPU
No dtype selected, switching to float64 to match model dtype.
MACE: v0 = 45.7100, e0 = -0.0468, B = 0.7390
EMT: v0 = 46.3859, e0 = -0.0062, B = 0.8378