Molecules
Molecules can be represented in various ways, each providing different levels of detail and information. In this section, we will explore some common representations of molecules, including molecular formulas, Lewis structures, SMILES notation. We will also introduce the concept of graphs and their relevance to chemistry and materials science.
SMILES¶
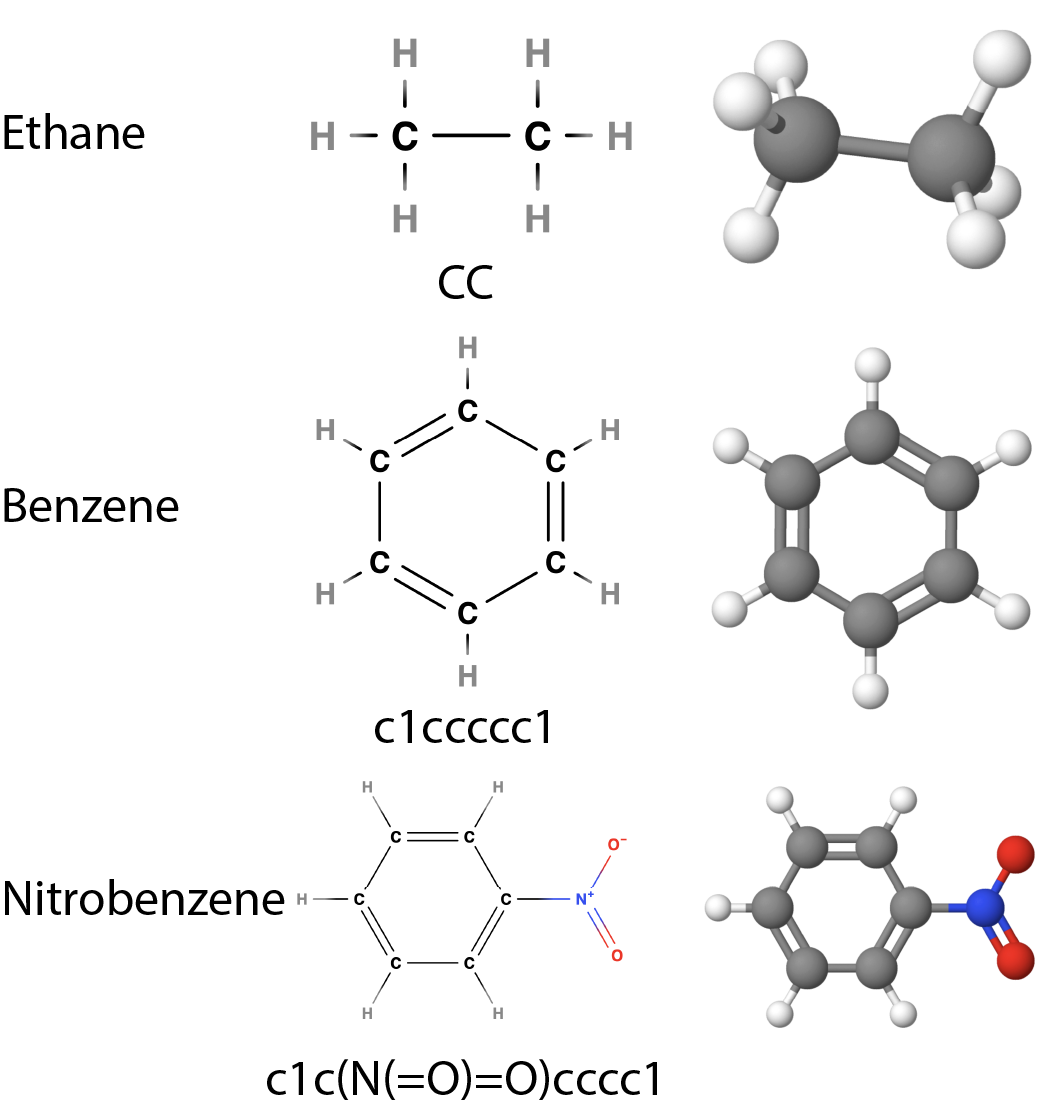
SMILES string representation of ethane, benzene, and nitrobenzene.
SMILES, which stands for Simplified Molecular Input Line Entry System, is a notation that allows the representation of a molecule’s structure using a line of text. This system is essential in cheminformatics because it translates complex molecular structures into a format that computers can easily process.
In SMILES notation, atoms are represented by their chemical symbols. For example, carbon is denoted by “C” and oxygen by “O”. Hydrogen atoms are usually implied and not explicitly written unless necessary for clarity, in which case they are enclosed in square brackets, such as [CH3].
Different types of chemical bonds are indicated using specific symbols. A single bond is typically represented by a dash (though it is often omitted as it is the default), a double bond by an equals sign “=”, a triple bond by a hash “#”, and an aromatic bond by a colon “:”.
To represent molecular structures, SMILES uses branches and rings. Branches are shown by enclosing the branch in parentheses, while ring structures are indicated by numbers that mark the start and end of the ring. Aromatic rings use lowercase letters for the atoms and are closed with numbers.
SMILES can also encode additional molecular information such as stereochemistry, charge, and even disconnected parts of a molecule. It is important to note that SMILES strings are not always unique; for example, ethane can be represented as “CC”, “C-C”, “[CH3]-C”, or “[CH3]-[CH3]”.
SMILES is widely used in fields such as chemistry and drug discovery. Tools like RDKit allow chemists and researchers to convert, manipulate, and analyze SMILES strings. This makes it easier to generate molecular structures from text, search chemical databases, and predict physical and chemical properties using computational models.
Overall, SMILES serves as a bridge between a molecule’s visual structure and its digital representation, making it a fundamental tool in modern chemical research and data analysis.
Graphs¶
A graph (G) is a mathematical structure used to model relationships between objects. It consists of vertices or nodes (V) and edges (E) that connect these nodes: G(V, E). Relationships in a graph are typically stored using a sparse adjacency matrix or list.
In the context of chemistry and materials science, graphs can be used to describe molecular structures. Here, atoms are represented as nodes and bonds as edges. Additional information, such as oxidation states, bond lengths, and bond strengths, can be stored as attributes of the nodes and edges.
Graphs are also valuable in machine learning. They can serve as inputs for algorithms, including Graph Neural Networks (GNNs), which we will discuss later in the course. GNNs can learn from the graph structure and node/edge attributes to make predictions or discover patterns.
By using graphs, we can effectively model and analyze complex relationships in molecular structures, making them a powerful tool in both theoretical and applied chemistry.